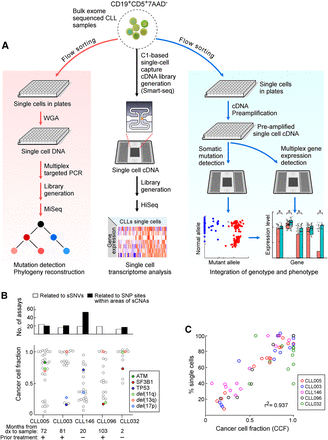
Detection of somatic alterations and gene expression patterns in single CLL cells. (A) Workflow of DNA and RNA analysis at the single-cell level. Viable leukemia cells from CLL patients were flow sorted either into 96-well plates or processed initially as bulk populations. DNA (left) and RNA (right) plate-based approaches were used for phylogeny reconstruction and integration of genotype and phenotype, respectively. Bulk cells (middle) were applied to C1 integrated fluidic circuits (IFCs) for single-cell capture and cDNA library generation (see Methods). Sequencing libraries were generated and sequenced on an Illumina HiSeq system. (B) Number of single-cell DNA-based detection assays designed (top) and the cancer cell fraction (CCF) of all the alterations (bottom panel) for five CLL samples. Each point is an alteration with specific alterations indicated by colors as noted. (C) Correlation between mutation and chromosomal abnormalities detected by single-cell DNA analysis and CCF inferred from bulk tumor whole-exome sequencing (WES).











