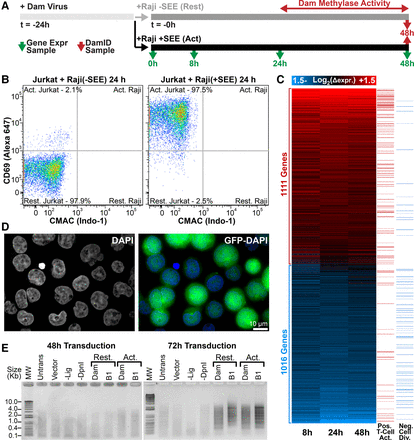
Transcription and genome organizational changes during T-cell activation. (A) Transcriptome and DamID experimental strategy. (B) FACS plot of CD69 versus CMAC staining for Jurkat T cells exposed to Raji B cells lacking/presenting SEE antigen. Raji B cells were gated from CMAC staining to distinguish them from Jurkat T cells, revealing 97.5% Jurkat T-cell activation. (C) Heatmap of time-course gene expression changes with a 1.4-fold cutoff highlighting GO terms positively associated with T-cell activation or negatively associated with cell division in the right-most columns (GO terms in Supplemental Fig. S1). (D) Representative micrograph of GFP-encoding lentivirus-transduced Jurkat T cells indicating high efficiencies for parallel transduced DamID constructs. (E) PCR amplified Dam-methylated genomic DNA. Amplification is only observed in resting (Rest.) or activated (Act.) cells 72 h post transduction. T4 DNA ligase-null (-Lig) and DpnI-null (-DpnI) controls represent background amplification. DamID results are derived from a single experiment, while gene expression analysis was performed in triplicate.











