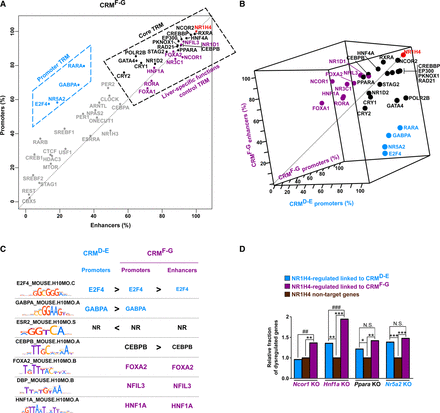
Main TRMs occurring at NR1H4-bound CRMs and functional validation of their role using dysregulated expression in the liver of mice KO for representative TRs. (A) Plot showing the occurrence of each TR at promoters versus enhancers from CRMsF-G. TRsshared, TRsD-E, and TRsF-G are depicted in black, blue, and violet according to Figure 2. Other TRs were displayed in gray. TRsshared and TRsF-G defining the core and liver-specific functions control TRMs on one hand and TRsD-E defining the promoter TRMs on the other hand are highlighted into dashed boxes. (B) Three-dimensional plot showing the occurrence of each TR at CRMD-E and CRMF-G promoters as well as CRMF-G enhancers. TRsshared, TRsD-E and TRsF-G are depicted in black, blue, and violet according to Figure 2. (C) DNA binding motifs enriched in CRMD-E and CRMF-G promoters and in CRMF-G enhancers (defined using regions from class A as controls) are indicated using the name of the recognizing transcription factor. Moreover, < and > were used to indicate significant differential enrichment within distinct sets of CRMs. (D) Fraction of NR1H4 target genes dysregulated in the liver of mice KO for the indicated TR. Genes exclusively associated with CRMsD-E or CRMsF-G and whose expression is modified in the liver of Nr1h4 KO mice were used for these analyses. Genes which are not linked to NR1H4-bound CRMs and whose expression is not altered in the liver of Nr1h4 KO mice (NR1H4 nontarget genes) served as the reference (arbitrarily set to 1). Fisher's exact test with Benjamini–Hochberg correction was used to define statistically significant differences with NR1H4 nontarget genes ([*] P < 0.05, [**] P < 0.01, and [***] P < 0.001) or between NR1H4-regulated genes linked to CRMsD-E and CRMsF-G ([##] P < 0.01, [###] P < 0.001, [N.S.] not significant).











