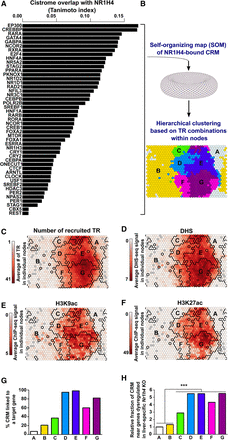
Integrative cistromics identifies the active subset of NR1H4-bound CRMs which consists of distinct classes of TRs recruiting CRMs. (A) Individual comparison of the NR1H4 cistrome in mouse liver with that of the 47 indicated TRs. (B) NR1H4-bound CRMs from the mouse liver genome were classified using a self-organizing map (SOM) based on their pattern of TR recruitment. Hierarchical clustering was subsequently used to identify seven main classes of CRMs which are indicated on the planar view of the toroidal map using different colors and denoted A to G. (C–F) The map issued from B was used to indicate the average number of binding TRs (C), the average DHS (D), H3K9ac (E), or H3K27ac (F) levels at CRMs contained in each node. Bold black lines indicate the borders of the clusters. (G) Percentage of CRMs from classes A–G potentially involved in gene transcriptional regulation. (H) Relative number of CRM from classes A–G found within 25 kilobases (kb) of the transcriptional start site (TSS) of genes whose expression is dysregulated in the liver of liver-specific Nr1h4 KO mice. This window allows the capture of a large fraction of distal sites able to influence gene expression (Akhtar et al. 2013). Fisher's exact test with Benjamini–Hochberg correction was used to define statistically significant differences between classes; (***) P < 0.001.











