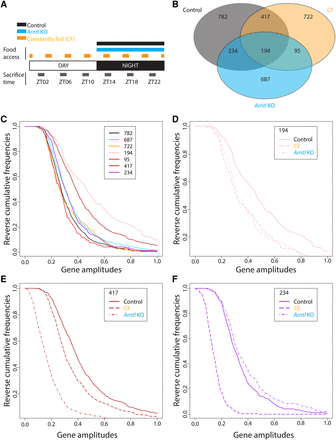
Rhythmic mRNAs in control, CF, and Arntl KO mouse liver. (A) Experimental design. Mice were fed over the course of 1 wk either only during the night (control and Arntl KO mice) or every 3 h (CF, constantly fed mice). They were then sacrificed every 4 h during two consecutive days (3–6 replicates per time point). For CF mice, the timing was established such that sacrifice always occurred 1 h after feeding. Liver RNA was extracted and used for gene expression microarray analyses. A cosine function was fitted to the data, and genes with a P-value associated with the fitting to the model lower than 0.0001 were considered as oscillating genes. (B) Venn diagram displaying the number of genes oscillating in each condition. The false discovery rates were 0.09 for WT mice, 0.08 for CF mice, and 0.09 for the Arntl KO mice (1000 permutations). (C) Reverse cumulative frequencies (RCF) of the amplitudes of the indicated group of genes (corresponding to the Venn diagram) in control mice (782, 417, 234, 194), CF (722, 95), and Arntl KO (687). RCF corresponds here to the frequency of all the values greater than a given amplitude. (D) RCF of the amplitude in the control, CF, and Arntl KO mice for the 194 genes oscillating in all the conditions. (E) As in D, but for the 417 genes oscillating in both control and CF mice. (F) As in D, but for the 234 genes oscillating in both the control and Arntl KO livers.











