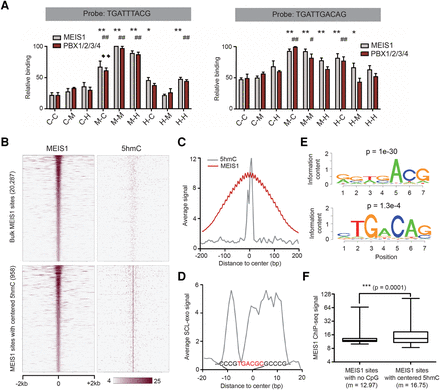
DNA binding by TALE-HD proteins is sensitive to cytosine modifications. (A) MEIS1 and PBX1/2/3/4 in vitro interaction with differentially modified synthetic PBX1/HOXA9 (left panel) and MEIS1/PBX1 (right panel) probes. Results are expressed according to the strongest binding equivalent to 100 (C: unmodified cytosine; M: methylated cytosine; H: hydroxymethylated cytosine; first letter: upper strand; second letter: lower strand). Statistical significance of differential binding to modified probes compared to the unmodified probe was tested with a Mann-Whitney test (n = 5; MEIS1 protein: [*] P < 0.05, [**] P < 0.01; PBX1/2/3/4: [#] P < 0.05, [##] P < 0.01). For PBX1 binding, ♦♦ corresponds to P < 0.01 between M-C and M-H conditions. (B) Heat maps of MEIS1 ChIP-seq and SCL-exo (5hmC) signals at 20,287 bulk MEIS1 binding sites (upper panels) and 958 MEIS1 sites showing a centered SCL-exo signal (lower panels) in P19 cell-derived NPCs. (C) Average MEIS1 ChIP-seq (red curve) and SCL-exo (gray curve) signals at the 958 MEIS1 binding sites showing a centered SCL-exo signal in P19 cell-derived NPCs. Note that oscillations in the MEIS1 profile are due to the lower resolution of the MEIS1 ChIP-seq .wig file compared to the SCL-exo .wig file. (D) Average SCL-exo signal over 40 bp for the cluster of 958 MEIS1 sites depicted in B. The consensus sequence determined from the aligned 958 sequences with the software CLC Sequence Viewer 6 is indicated on the graph. A motif related to the TGACAG consensus MEIS1 motif has been highlighted in red font. (E) Examples of de novo motifs found enriched (P-value is indicated above the logos) by the SeqPos motif tool from Cistrome. (F) Box plot analysis of MEIS1 ChIP-seq signal for MEIS1 sites with no CpGs within the ChIP-seq peaks (n = 3628) and the 958 MEIS1 sites with centered SCL-exo signal (m = mean ChIP-seq signal).











