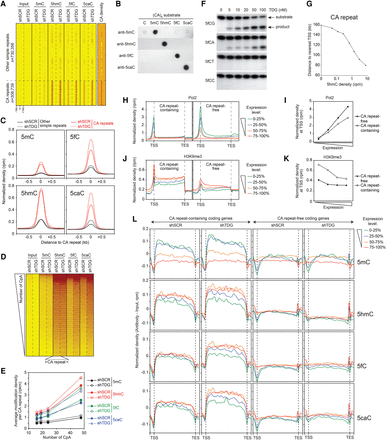
DNA methylation dynamics at CA repeats. (A) Heatmaps of 5mC/5hmC/5fC/5caC/CA densities in control and Tdg-deficient MEFs at simple repeats sorted by family. Note the specific enrichment of 5mC oxidation derivatives at CA repeats. Tags were counted within 1 kb around the simple repeat center. (B) Anti-5mC, anti-5hmC, anti-5fC, and anti-5caC antibodies do not recognize CA repeats nonspecifically. Dot blot assays showing that 5mC, 5hmC, 5fC, and 5caC antibodies specifically recognize 5mC, 5hmC, 5fC, and 5caC-containing substrates, respectively in (CA)9 repeat contexts. (C) Average 5mC/5hmC/5fC/5caC signals at CA repeats reveal a specific accumulation of 5fC and 5caC in the absence of TDG. (D) Heatmaps of 5mC/5hmC/5fC/5caC levels in control and Tdg-deficient MEFs at CA repeats ranked in descending order based on their number of CpA dinucleotides. (E) Curves showing the positive correlation between cytosine modification densities and CA densities at CA repeats. CA repeats were sorted in quartiles based on their number of CpA dinucleotides. (F) In vitro glycosylase assays revealing that the recombinant protein TDG excises formylcytosine exclusively in a CpG or CpA context. (G) Average distance to TSS of CA repeats as a function of their 5hmC level. (H–K) Normalized densities of Pol2 (H,I) and H3K9me3 (J,K) within gene bodies (H,J) or at TSSs (I,K) expressed at different levels. Genes were sorted into two groups according to the presence or the absence of CA repeats (length >100 bp) within their gene body (CA repeat-containing and CA repeat-free, respectively). Tag densities were collected in 100-bp sliding windows spanning 2 kb (divided in 10 bins) of the length-normalized gene bodies (divided in 40 bins). Within both groups, genes were then sorted in quartiles according to their expression level. (L) Average 5mC/5hmC/5fC/5caC signals in control and Tdg-deficient MEFs within CA repeat-containing and CA repeat-free gene bodies expressed at different levels. Tag densities were collected in 100-bp sliding windows spanning 2 kb (divided in five bins) of the length-normalized gene bodies (divided in 50 bins).











