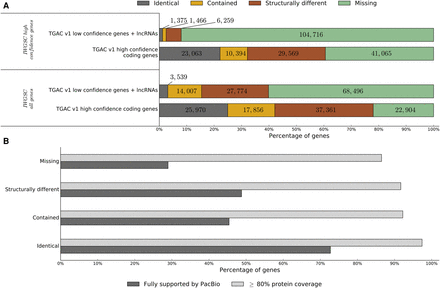
Comparison between IWGSC annotation and TGACv1 high (HC) and low confidence (LC) genes. IWGSC genes were aligned to the TGACv1 assembly (gmap, ≥90% coverage, ≥95% identity) and classified based on overlap with TGACv1 genes. (A) Identical indicates shared exon–intron structure; contained, exactly contained within the TGACv1 gene; structurally different, alternative exon–intron structure; and missing, no overlap with IWGSC. (B) Bar plot showing proportion of HC TGACv1 protein-coding genes supported by protein similarity or PacBio data. Genes are classified based on overlap with the full set of IWGSC genes.











