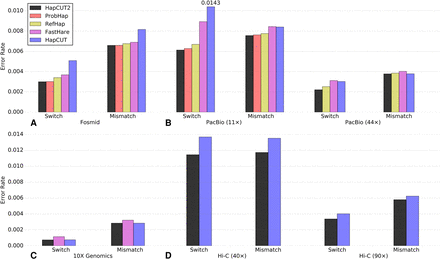
Accuracy of HapCUT2 compared to four other methods for haplotype assembly on diverse whole-genome sequence data sets for NA12878. (A) Fosmid dilution pool data (Duitama et al. 2012). (B) PacBio SMRT data (11× and 44× coverage). (C) 10X Genomics linked reads. (D) Whole-genome Hi-C data (40× and 90× coverage, created with MboI enzyme). Switch and mismatch error rates were calculated across all chromosomes using the subset of variants that were phased by all methods. For each data set, only methods that produced results within 20 CPU-h per chromosome are shown.











