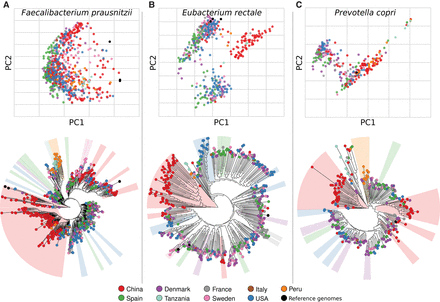
Population genetic structure of three common intestinal species and its association with sampling geography. Strain population structures for three representative human gut species, reported both as phylogenies built on the concatenated alignments of each species-specific reconstructed marker set (bottom). To highlight the presence of discrete clusters of related strains, we also report the genetic distances measured on the alignments as principal coordinate ordinations (top). We report the population structure of Faecalibacterium prausnitzii (A), Eubacterium rectale (B), and Prevotella copri (C). Results for additional species are reported in Supplemental Figures S12–S16, S18–S24.











