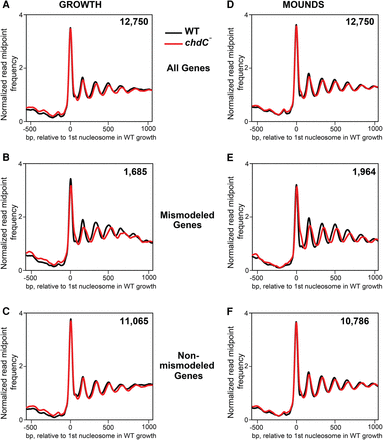
Nucleosomes are mismodeled in a subset of genes during growth and development of chdC-null cells. (A) Normalized read midpoint frequencies in WT (black) and chdC-null (red) growing cells for all 12,750 genes aligned to the midpoint of the first defined nucleosome in WT growing cells. (B) Normalized read midpoint frequencies in WT (black) and chdC-null (red) growing cells for the 1685 mismodeled genes in growing chdC-null cells aligned to the midpoint of the first defined nucleosome in WT growing cells. (C) Normalized read midpoint frequencies in WT (black) and chdC-null (red) growing cells for the 11,065 non-mismodeled genes in growing chdC-null cells aligned to the midpoint of the first defined nucleosome in WT growing cells. (D) Normalized read midpoint frequencies in WT (black) and chdC-null (red) loose-mound–stage cells for all 12,750 genes aligned to the midpoint of the first defined nucleosome in WT growing cells. (E) Normalized read midpoint frequencies in WT (black) and chdC-null (red) loose-mound–stage cells for the 1964 mismodeled genes in mound-stage chdC-null cells aligned to the midpoint of the first defined nucleosome in WT growing cells. (F) Normalized read midpoint frequencies in WT (black) and chdC-null (red) loose-mound–stage cells for the 10,786 non-mismodeled genes in loose-mound–stage chdC-null cells aligned to the midpoint of the first defined nucleosome in WT growing cells.











