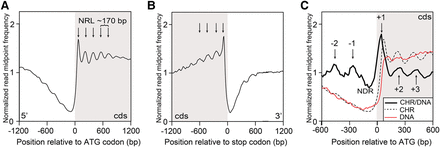
Genome-wide nucleosome positioning in Dictyostelium. (A) Normalized read midpoint frequency distributions of MNase-protected fragments (nucleosome dyads) of all 12,750 genes in growth-stage WT cells were aligned relative to their ATG codons. Peaks (arrows) correspond to dyad midpoints for globally phased nucleosomes in the 5′ region of intragenic DNA, and distances between mapped read peaks correspond to ∼170 bp NRL. The protein coding DNA sequence (cds) region is shaded. (B) Normalized read midpoint frequency distributions of all genes in growth-stage WT cells were aligned relative to their translational termination sites (stop codons). Peaks (arrows) in the mean normalized frequency distribution correspond to globally phased nucleosomes in the 3′ region of intragenic DNA. The protein cds region is shaded. (C) Normalized dyad read midpoint frequency distributions for WT chromatin (CHR; dotted line) (see A) were adjusted for sequence mappability by dividing with equivalent control data from MNase-digested naked (protein free) WT DNA (DNA; red line) and replotted as the ratio (CHR/DNA; thick black line) within 1.2 kb of flanking chromatin relative to ATG sites of all 12,750 genes. An ∼170-bp nucleosome-depleted (“free”) region (NDR) is centered near the AT-rich regions of Dictyostelium TSS. Positioned nucleosomes upstream (+) and downstream (−) to the NDR are indicated by arrows. The protein cds region is shaded.











