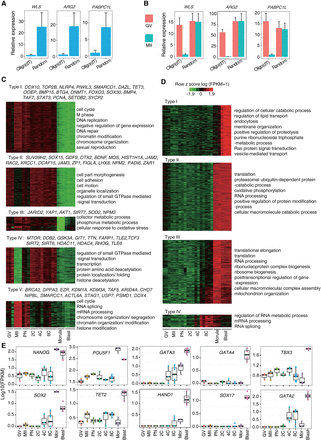
Classification of maternal and zygotic genes. (A) Real-time PCR examination in cDNA samples of MII oocytes. Gene expression levels were significantly higher in samples reverse-transcribed with random primers than in those prepared with oligo(dT) primers. (B) Real-time PCR examination in cDNA samples of GV and MII oocytes. Gene expression levels were comparable between GV and MII oocytes when cDNAs were prepared with random primers, but showed significant difference when cDNAs were prepared with oligo(dT) primers. (C) Five expression patterns of maternal genes were identified. Typical genes and representative GO terms are listed. (D) Four expression patterns of zygotic genes were identified. Their representative GO terms are shown. (E) The expression patterns of several zygotic genes implicated in the cell-fate determination. (C–E) Based on monkey individual-oocyte/embryo sequencing data. Similar results for the pooled-embryo data are shown in Supplemental Figures S4 and S5.











