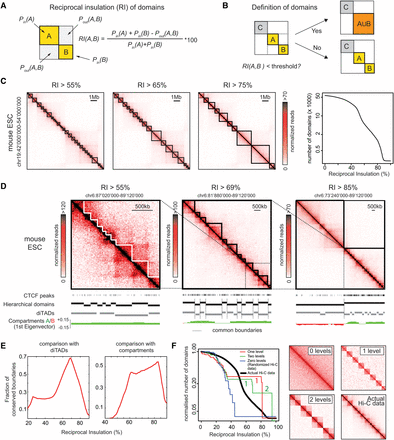
Schematic description of reciprocal insulation and the domain-calling algorithm. (A) Schematic representation of reciprocal insulation (RI) between two fictitious domains A and B in Hi-C data. (B) The CaTCH algorithm merges two adjacent domains if their reciprocal insulation is smaller than a given threshold. (C) (Left three panels) Examples of sets of domains defined in mouse ESCs Hi-C data (20-kb binning) imposing different threshold on RI. (Right) Number of domains detected in ESC as a function of RI. (D) Sub-TAD contact domains (left), directionality index-based TADs (middle), and A/B compartments (right) are identified at different RI values. (E) Fraction of boundaries of diTAD (left) and compartments (right) overlapping with boundaries of domains identified by CaTCH as a function of RI. (F) (Left) Number of domains detected by CaTCH as a function of RI in the real genome (black line), or in computationally generated contact maps with zero (blue), one (red), or two preferential folding levels (green). The corresponding heat maps are shown in the four right panels. Numbers of domains were normalized to the initial step (0% insulation) to allow comparison.











