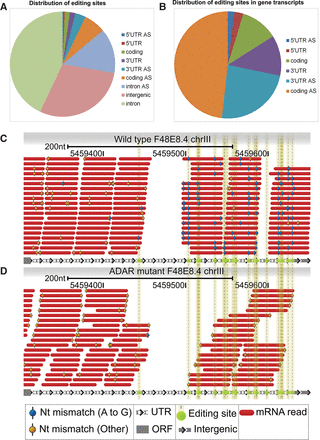
Most of the editing sites in nonrepetitive regions are located in intergenic and intronic regions or at 3′ UTR in gene transcripts. (A) Pie graph presenting the distribution of editing sites that were found by nonrepetitive alignment to the genome. Except for sites in intergenic regions that are not assigned to genes, all other sites were annotated as either sense or antisense (AS) to the assigned gene. (B) Distribution of editing sites located in gene transcripts, 5′ UTR, coding, or 3′ UTR regions. Sites were separated based on the region and orientation to the gene, sense, or antisense (AS). (C,D) Visualization of editing sites found at the 3′ UTR of the F48E8.4 gene. 3′ UTR of the gene is presented by arrows. (C) Sequences from wild-type worms. (D) Sequences from ADAR mutant worms. Red bars represent reads aligned to the 3′ UTR of F48E8.4 by nonrepetitive alignment from one library used in this study. Yellow lines are the predicted editing sites by this analysis. Blue dots on the sequences show A-to-G nucleotide changes, and orange dots show other nucleotide changes found in the sequences. Regions that do not have sequence coverage are repetitive regions. Therefore, sequences that aligned to them also aligned to other regions in the genome and were excluded from the analysis. The general abundance of A-to-G mismatches in the edited area can be observed in the wild type, in contrast to the complete lack of such changes in the ADAR mutant.











