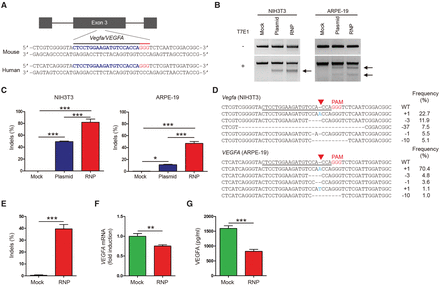
Targeted mutagenesis in the Vegfa/VEGFA gene via Cas9 ribonucleoproteins (RNPs). (A) The target sequence in the Vegfa/VEGFA locus. The PAM sequence and the sgRNA target sequence are shown in red and blue, respectively. (B) Vegfa-specific Cas9 RNP-driven mutations in NIH3T3 and ARPE-19 cells detected by the T7 endonuclease I (T7E1) assay. Arrows indicate the expected positions of DNA bands cleaved by T7E1. (C) Mutation frequencies measured by targeted deep sequencing. Error bars indicate SEM (n = 3). One-way ANOVA and Tukey post-hoc tests: (*) P < 0.05; (***) P < 0.001. (D) Representative mutant DNA sequences at the Vegfa/VEGFA locus. The PAM sequence is shown in red, and inserted nucleotides are shown in blue. The target sequence is underlined. The red triangle indicates the cleavage position. The column on the right indicates the number of inserted or deleted bases and indel frequencies (%). (WT) wild type. (E) Vegfa-specific Cas9 RNP-driven mutation frequencies in confluent ARPE-19 cells at 64 h post-transfection detected by targeted deep sequencing. (F) Relative VEGFA mRNA levels measured by quantitative PCR (qPCR). (G) Secreted VEGFA protein in the supernatant measured by ELISA. Error bars indicate SEM (n = 5). Student's t-test: (**) P < 0.01; (***) P < 0.001.











