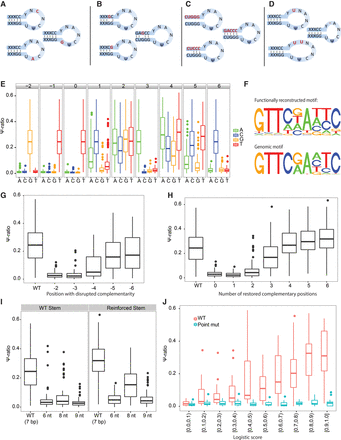
Validation of TRUB1 consensus motif using MPRA analysis. (A–D) Scheme of systematic mutations employed in this study, perturbing the sequence of the loop (A), individual positions in the stem structure (B), all positions in the stem structure along with compensatory mutations systematically restoring complementarity (C), and the size of the loop (D). (E) Seventy-four sites containing a TRUB1 consensus motif were systematically point-mutated at each position. Boxplots capturing the distribution of Ψ-ratios across each of the 74 sites are depicted in each of the perturbations. (F) (Top panel) For each of the indicated positions, we first extracted the median Ψ-ratio obtained using each of the 4 nt. The median Ψ-ratio for each of these nucleotides was then divided by the sum of the Ψ-ratios across all 4 nt, to yield relative Ψ-ratios (summing up to 1, at each position). The height of each nucleotide at each position was then plotted in direct proportion to its relative Ψ-ratio. (Bottom panel) The sequence motif of TRUB1, as identified in Figure 1D, is plotted to ease the comparison with the functionally defined motif. (G) Distribution of Ψ-ratios, following disruption of the base-pairing ability of each of the indicated positions in the stem structure. The distribution for WT sequences is presented in comparison. (H) Distribution of Ψ-ratios following elimination and gradual sequential restoration of the stem structure, beginning with zero complementary bases up to six complementary bases. (I) Distribution of Ψ-ratios following extension (to 8 or 9 nt) or shrinking (to 6 nt) of loop length, either based on the WT TRUB1 sites (left), or based on variants with particularly strong stems (right) of six consecutive base pairs. (J) Distribution of Ψ-ratios across 250 sites with varying logistic scores of TRUB1-mediated pseudouridylation. Twenty-five sites were selected for each of 10 logistic score bins, ranging from 0 to 1. As controls, distributions of Ψ-ratios are shown also for 250 counterparts in which we designed a T→C point-mutation at the pseudouridylated site.











