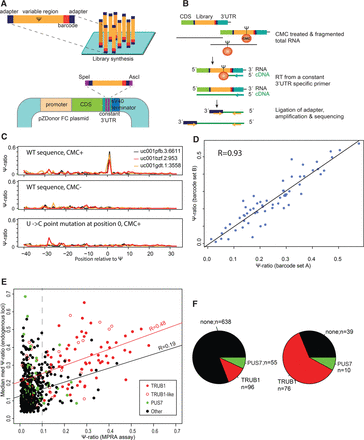
Establishment of massively parallel reporter assay and validation of selected targets. (A) Thousands of sequence variants surrounding pseudouridylated sites or mutated counterparts, each harboring a unique 8-nt barcode and flanked by an adapter set, are cloned downstream from a reporter gene and transfected into cells. (B) Strategy employed for obtaining targeted readouts of pseudouridine within the constructs. Following CMC-treatment, total RNA was reverse-transcribed using a construct-specific primer. A DNA adapter was subsequently ligated to the cDNA, and DNA was amplified using one primer harboring complementarity to the adapter sequence and a second one downstream from the sequence employed for reverse-transcription (Methods). (C) Ψ-ratios across a 70-nt window surrounding three endogenously pseudouridylated sites at TRUB1 targets, within the indicated genes. In all three cases, the pseudouridylation is precisely recapitulated at the correct site in the WT, CMC-treated sample (upper panel) but completely eliminated in the absence of CMC treatment (middle panel), or upon point-mutation of the pseudouridylated site (lower panel). (D) Scatterplot presenting the correlation between Ψ-ratios measured for identical sequences (the set of 74 WT TRUB1 sites), differing only in their 8-nt barcode. (E) Correlation between Ψ-ratios, as captured in the massively parallel reporter assay, and the median med-Ψ- ratios measured across the three large data sets analyzed in this study. TRUB1 sites are defined as harboring a GTTCNANNC consensus, and TRUB1-like sites are defined as GTT[A/G/T]NANNC. The regression curve is plotted in red for all TRUB1 and TRUB1-like sites, in black for all remaining sites. (F) Pie chart depicting the distribution of TRUB1, PUS7, and other consensus sequences throughout the 789 validated sites (left panel) and among all sites with Ψ-ratios > 0.1 (right panel).











