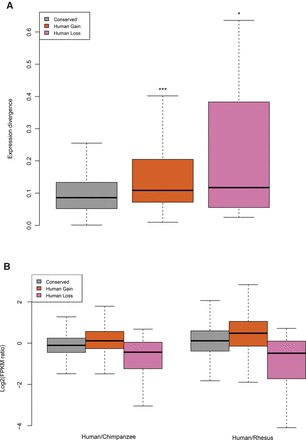
m6A evolution and gene expression divergence. (A) The expression divergence of m6A-modified orthologous genes were plotted along the y-axis. These values were calculated from the coefficient of variation of log2 transformed individual FPKMs. The Wilcoxon test was performed to calculate significant level of statistical differences between “conserved” and the other groups: (***) P < 10−8, (*) P < 10−2. (B) The expression change of orthologous genes was plotted against groups of m6A-modified genes. The “conserved” group has a similar number of up- and down-regulated genes. Human gain m6A-modified genes demonstrated more genes up-regulated compared with chimpanzee and rhesus orthologs, while human loss genes showed more genes were down-regulated. (Left) Human compared to chimpanzee; (right) human compared to rhesus. Conserved, N = 2118; human gain, N = 250; and human loss, N = 30.











