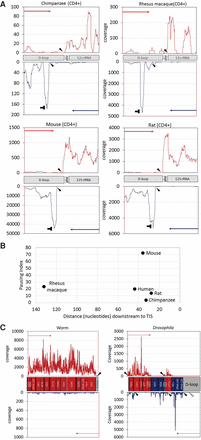
Identification of mtDNA nascent transcript across evolution: (A) PRO-seq experiment performed in four mammalian CD4+ cells: chimpanzee, rhesus macaque, rat, and mouse. x- and y-axes are identical to those in Figure 2, and “horizontal T” sign designates the pausing site. Filled arrowheads in all panels point to the calculated identified TIS. Notably, in three species (chimpanzee, rat, and mouse), the major heavy-strand TIS was identified downstream from the tRNA phenylalanine gene, similar to the human heavy-strand TIS 1. (B) Pausing site of light-strand transcription in mammals. (x-axis) Distance (in nucleotides) of the pausing site from the light-strand TIS; (y-axis) pausing index value of each species. The name of each species is indicated to the right of each dot. (C, left) Analysis of GRO-seq data from worm. In this species, there is a single TIS for a single transcription unit, present only at the heavy strand. (C, right) Analysis of PRO-seq data from Drosophila. Five candidate TISs were identified: two in the heavy strand and three in the light strand. Two minor additional TISs are marked by empty arrowheads.











