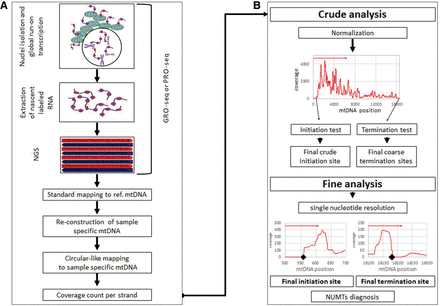
Workflow of analysis. (A) GRO-seq and PRO-seq experiments generate genome-wide nascent transcript data. The extracted mtDNA sequences enable reconstruction of sample-specific mtDNA sequence, which is used in turn as a circular-like mapping reference. This allows counting the sequencing read coverage in a strand-specific manner. (B) Analysis of mtDNA transcription initiation and termination sites. Two steps were designed: (1) a crude step for candidate transcription initiation site (TIS) identification, identifying abrupt increase in read coverage in a nucleotide resolution within 200 bp sliding windows; and (2) fine analysis, focusing on highest scoring regions to identify the best TIS candidate. Notably, the identification of transcription termination sites utilizes the same approach, yet instead of an abrupt increase in reads, an abrupt decrease in read coverage is identified.











