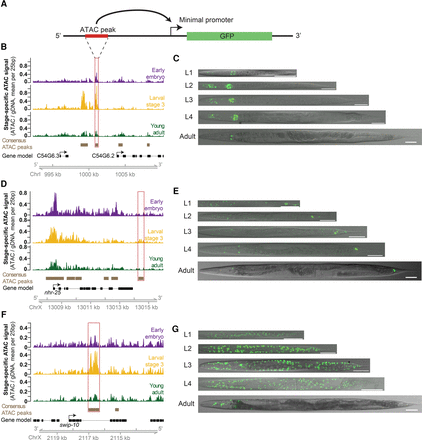
Dynamic ATAC-seq peaks identify functional enhancers with unique spatiotemporal specificity during C. elegans development. (A) Functional enhancer constructs used to generate C. elegans transgenic lines. Putative regulatory regions, or corresponding flanking regions for negative controls, are inserted upstream of a minimal promoter (pes-10) driving green fluorescent protein (GFP) localized to the nucleolus. (B,D,F) Distal (>1 kb from a TSS) noncoding ATAC-seq peaks with the largest fold change in ATAC-seq signal between any two stages were screened for potential enhancer activity. The approximate regions tested near C54G6.2 (B), nhr-25 (D), and swip-10 (F) are boxed in red. Note that for swip-10 (F), the plot orientation is reversed for consistency with other plots. (C,E,G) Specific patterns of spatiotemporal enhancer activity in transgenic lines. Representative images of GFP expression in staged C. elegans transgenic lines are presented with a 50-µm scale bar. All images were straightened with ImageJ and are grayscale images with florescence overlaid.











