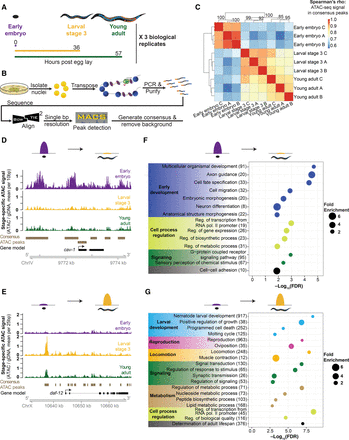
ATAC-seq in whole C. elegans captures chromatin accessibility dynamics across three life-stages. (A) Three independent biological replicates each consisting of tightly temporally synchronized C. elegans were used for ATAC-seq. Hours post-egg lay are at 20°C. (B) C. elegans were flash frozen and nuclei were isolated before assaying accessible chromatin using transposons loaded with next-generation sequencing adaptors, allowing paired-end sequencing. A custom analysis pipeline emphasizing high-resolution signal and consistent peaks, as well as accommodating input control, was developed to generate stage-specific and consensus (i.e., across stages) ATAC-seq peaks. (C) ATAC-seq signal within consensus ATAC-seq peaks was compared between all samples using Spearman's ρ to cluster samples. Replicate batches are noted as letters following the stage. (D,E) Comparison of ATAC-seq signal (normalized by total sequencing depth) between all three stages at a region that decreases (D) or increases (E) in accessibility during development. (F,G) Genes that lose accessibility between embryo and larval stage 3 (L3) are enriched for early development functions (F), while genes that gain accessibility are enriched for larval-related functions (G); all calculations and genes lists are from GOrilla (Eden et al. 2009), and the number of genes enriched in each term are listed in parentheses.











