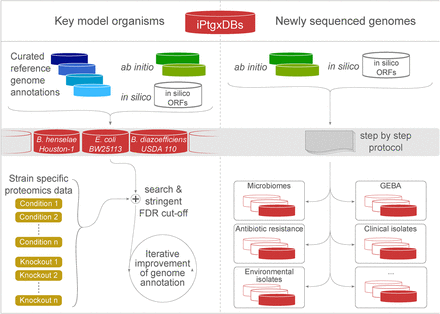
Application of our integrated proteogenomics approach. We release open source iPtgxDBs for several model organisms (https://iptgxdb.expasy.org); here, for Bhen, E. coli BW25113, and B. diazoefficiens USDA 110 (left panel). Using proteomics data from any condition or knockout strain (light brown boxes; here, schematically shown for E. coli), researchers can identify novelties and iteratively improve the genome annotation, e.g., in a community-driven genome Wiki approach (Salzberg 2007). The release of the software to integrate ab initio predictor(s) and in silico predictions (Supplemental Fig. S8) can help to improve genome annotations of many newly sequenced genomes (right panel).











