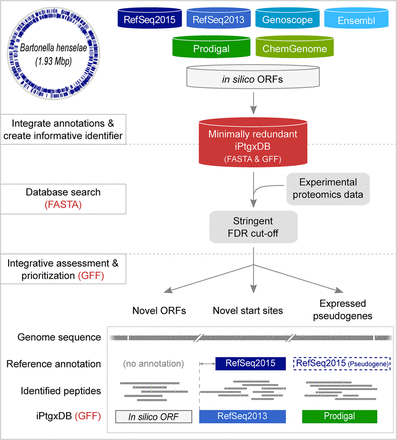
Integrative proteogenomics workflow. For a completely sequenced prokaryotic genome (Bhen is shown as an example with annotated CDSs), reference genome annotations (blue containers), results from ab initio gene prediction algorithms (green containers), and in silico ORFs (white container) are downloaded or computed and integrated in a first preprocessing step (upper panel). All CDS and pseudogene annotations are matched, and informative gene identifiers are created and stored in a minimally redundant iPtgxDB (red container; searchable protein sequences in FASTA format, integrated annotations in GFF format). Experimental proteomics data are matched to the DB using a target-decoy approach relying on stringent FDR cut-offs (middle panel). Identified PSMs and peptides are postprocessed to visualize novel candidates (lower panel) in the context of experimental data integrated with the GFF file.











