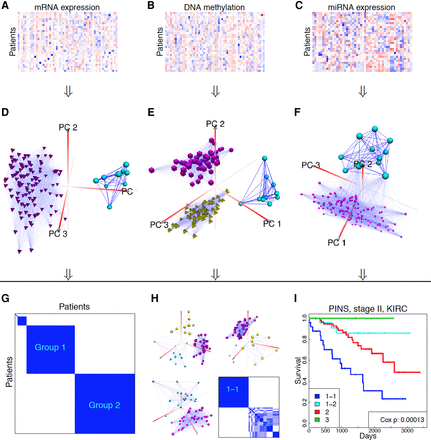
Data integration and disease subtyping illustrated on the kidney renal clear cell carcinoma (KIRC) data set. (A–C) The input consists of three matrices that have the same set of patients but different sets of measurements. (D–F) The optimal connectivity between the samples for each data type. (G) The similarity between patients that are consistent across all data types. Partitioning this matrix results in three groups of patients. (H) Group 1 is further split into two subgroups in stage II. (I) Kaplan-Meier survival curves of four subtypes after stage II splitting of group 1. The survival analysis indicates that the four groups discovered after stage II have significantly different survival profiles (Cox P-value 0.00013).











