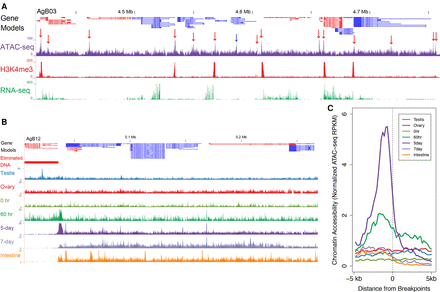
Increased chromatin accessibility is associated with Ascaris CBRs. (A) ATAC-seq identifies accessible chromatin at Ascaris promoters. Gene models, transcripts (red, plus strand; blue, minus strand) and H3K4me3 ChIP-seq illustrated in a genome region of 5-d embryos (32- to 64-cell stage). For ATAC-seq data, red arrows indicate peaks of more accessible chromatin near transcriptional start sites, while the blue arrow points to open chromatin within a gene. Note the ATAC-seq signal is enriched at promoters/transcription start sites. Also illustrated are RNA-seq data for 5-d embryos. (B) Ascaris CBRs exhibit increased chromatin accessibility just prior to DNA elimination. The developmental ATAC-seq profile for a DNA breakpoint region (from AgB03) is illustrated. Note that in addition to the transcription start site–associated ATAC-seq peaks, a broad area of open chromatin appears within the break region at the four-cell stage (60 h) immediately prior to DNA elimination. This region remains open through early development (5 d; gastrulation) but is closed in late embryos (7 d; morphogenesis) and somatic tissues (intestine and muscle). (C) Chromatin accessibility for 40 Ascaris breakpoint regions. Illustrated are the breakpoints with their 5-kb flanking regions. Upstream of the breakpoints (−5 kb to 0) are the retained DNA, while downstream regions (0 to 5 kb) are the eliminated regions. Note the open chromatin at the breakpoints in 60 h (immediately prior to DNA elimination) that increases and persists in 5-d embryos. In addition, note that the open regions correspond exactly to the CBRs defined by chromosomal breaks and telomere addition.











