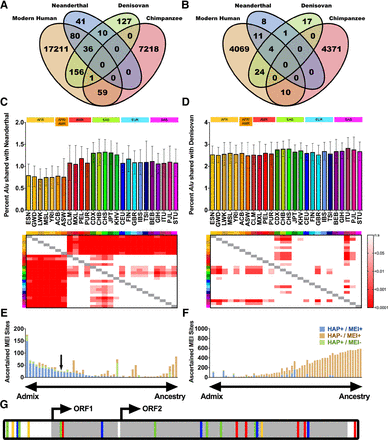
Mobile element activity in ancient human genomes and introgression of ancient MEIs into modern humans. (A,B) Sharing of (A) Alu and (B) L1 MEIs between Neanderthal, Denisovan, modern humans, and chimpanzees (Supplemental Table S11). (C,D) Sharing of Neanderthal and Denisovan Alu MEIs in each of the 26 1000 Genomes Project Phase III populations. For each population, we determined the average percentage per individual of Alu MEIs shared with (C) Neanderthal or (D) Denisovan. Heat maps represent multiple comparison ANOVA P-values between each population (key at right). (E) Analysis of Neanderthal MEI introgression in non-African individuals. Each bar represents one MEI site that was shared between Neanderthal and non-African individuals (i.e., the site was found only in SAS, EUR, and/or EAS). Bars are colored by MEI overlap with Neanderthal haplotypes (Supplemental Methods; Sankararaman et al. 2014), with sites to the left of the chart likely contributed to modern humans by introgression from Neanderthals and sites to the right likely due to common ancestry. “HAP” indicates whether the Neanderthal haplotype is present at the site (HAP+ or HAP−). “MEI” indicates whether the MEI is present at the site (MEI+ or MEI−). Blue bars indicate a high degree of linkage disequilibrium (LD) between the Neanderthal haplotype and the MEI (HAP+/MEI+), and brown bars indicate little or no correlation between the Neanderthal haplotype and the MEI (HAP-/MEI+). Blue sites have r2 values for HAP+/MEI+ of >0.5, whereas brown sites segregate independently (Supplemental Table S11; Supplemental Methods). The black arrow indicates the FL-L1 element described in G. (F) Analysis of Neanderthal MEI introgression in all individuals. Identical analysis to E but for sites with an AFR allele frequency greater than zero. (G) Cartoon of an FL-L1 element sequenced from a GBR individual, with differences shown as in Figure 3F. The quality of ancient MEI calls was comparable to those called in modern humans (Supplemental Fig. S12).











