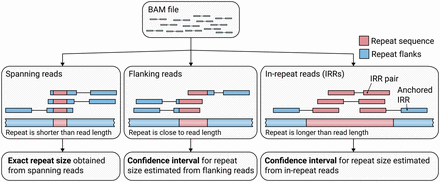
An outline of how ExpansionHunter catalogs reads associated with the repeat locus of interest and estimates repeat lengths starting from a binary alignment/map (BAM) file. (Left) Exact sizes of short repeats are identified from spanning reads that completely contain the repeat sequence. (Middle) When the repeat length is close to the read length, the size of the repeat is approximated from the flanking reads that partially overlap the repeat and one of the repeat flanks. (Right) If the repeat is longer than the read length, its size is estimated from reads completely contained inside the repeat (in-repeat reads). In-repeat reads anchored by their mate to the repeat region are used to estimate the size of the repeat up to the fragment length. When there is no evidence of long repeats with the same repeat motif elsewhere in the genome, pairs of in-repeat reads can also be used to estimate the size of long (greater-than-fragment-length) repeats.











