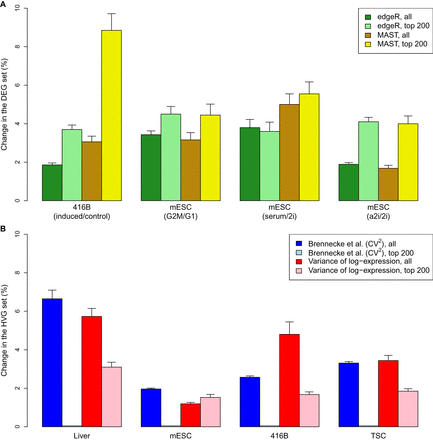
Effect of spike-in variability on DEG or HVG detection in simulated data. (A) The percentage change in the set of DEGs detected in each data set at a FDR of 5% by edgeR or MAST. This was also calculated for the top set of 200 DEGs with the smallest P-values. Simulations were performed to detect DEGs in our 416B data set after inducing expression of a CBFB-MYH11 oncogene compared to an mCherry control (Methods), between mouse embryonic stem cells (mESCs) in G1 and G2/M phases of the cell cycle (Buettner et al. 2015), or between mESCs cultured in different conditions—serum, ground state (2i), or alternative ground state (a2i) (Grün et al. 2014; Kolodziejczyk et al. 2015). (B) The percentage change in the set of HVGs detected in each data set at a FDR of 5%, using the method of Brennecke et al. (2013) based on the squared coefficient of variation (CV2) or with a method based on the variance of log-expression. This was also calculated for the top set of 200 HVGs with the smallest P-values. Simulations were performed to detect HVGs in our 416B and TSC data sets, in liver cells (Scialdone et al. 2015), and in mESCs (Kolodziejczyk et al. 2015). All values represent the mean of 20 simulation iterations, and error bars represent standard errors.











