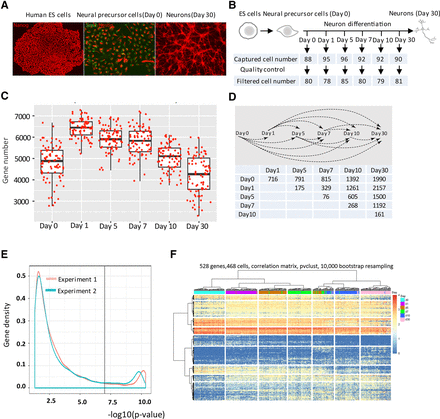
Transcriptional profiling and statistical properties of gene expression data of the neuronal differentiation process. (A) Immunostaining shows expression of markers for ES cells (NANOG), NPC (SOX2/NESTIN), and neurons (TUJ1). (B) Summary of workflow for capture and quality assessment of single cells at each time point. (C) Box plots show the number of expressed genes for each filtered cell at days 0, 1, 5, 7, 10, and 30 during neuron differentiation. Each dot represents one cell. (D) Summary of 15 pairwise comparisons (upper) that identified 3986 DE genes between any two of six time points at P < 0.05 (Supplemental Table S3). The table shows the numbers of DE genes for each pairwise comparison. (E) After comparing any two time points as shown in D, to enhance pattern detection of DE genes for downstream analysis, curves were drawn based on the gene density and −log10 (P-value) for all DE genes in experiment 1 (orange line) and experiment 2 (green line), respectively (E), from which, 528 genes were obtained (Supplemental Table S4) with fold change >1.5 and P-value <10−7. (F) Hierarchical cluster analysis for all six time points was performed by bootstrapping based on the derived 528 genes from E. Hierarchical cluster analysis was calculated for the cells clustering with the R package “pvclust” using Correlation distance, the Ward clustering method, and the number of bootstrap set to 10,000.











