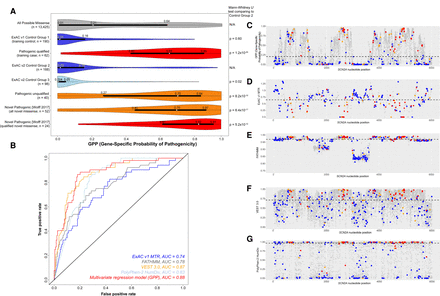
Real-time validation of a SCN2A gene-specific model. (A) SCN2A gene distributions of the GPP scores. All Mann-Whitney U tests compare groups to ExAC v2 Control Group 2. Control Groups 1–3 are mutually exclusive presumed benign missense variants. Pathogenic qualified, unqualified, and novel are mutually exclusive presumed pathogenic missense variants. For the bottom two plots of novel variants in Wolff et al. (2017), the “qualified novel” group is a “de novo” and severe pediatric epilepsy subset of the ‘all novel’ group. (B) ROC curves for the model and individual features accurately predicting the 52 novel case and 188 Control Group 2 variants. (C–G) Distribution of the model and individual feature scores across all 13,425 possible SCN2A missense variants (gray) with the median SCN2A score depicted by a dashed black line. Also plotted are the 188 ExAC v2 Control Group 2 (blue), the 52 novel variants from Wolff et al. (2017) (red), and the 40 SCN2A unqualified pathogenic test variants (orange).











