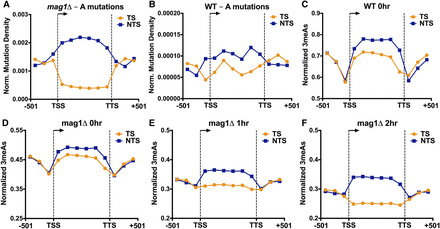
MMS-induced A-mutations are enriched on the nontranscribed strand (NTS) and depleted from the transcribed strand (TS) of yeast genes, particularly in a BER-deficient (mag1Δ) strain. (A,B) The density of mutations of “A” nucleotides in the MMS-treated (A) mag1Δ and (B) WT strains was plotted for 5762 yeast genes. The number of A-mutations in the transcribed strand (TS; orange) and nontranscribed strand (NTS; blue) was determined for bins spanning 501 bp upstream of the transcription start site (TSS), the transcribed region of the gene, and 501 bp downstream from the transcription termination site (TTS), and normalized by the number of A nucleotides in each bin. (C,D) 3-methyladenine (3meA) lesions are elevated on the NTS immediately following MMS-treatment (i.e., 0 h) in (C) WT and (D) mag1Δ strains. The average number of 3meA lesions per A nucleotide was plotted for bins spanning 5762 yeast genes and flanking promoter and downstream DNA sequences, as described in part A. (E,F) Levels of 3meA lesions are preferentially depleted from the TS in a mag1Δ strain during a repair time course. The average number of 3meA lesions per A nucleotide in the mag1Δ (E) 1-h and (F) 2-h time points was plotted as described in part C.











