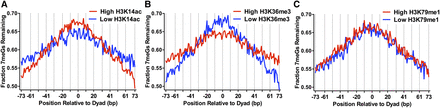
Histone post-translational modifications (PTMs) alter 7meG repair within nucleosomes. (A) Fraction of 7meG lesions remaining following 2-h repair in WT relative to 0-h mag1Δ control among top 10,000 nucleosomes with highest levels of H3K14 acetylation (High H3K14ac) and 10,000 nucleosomes with the lowest levels of H3K14 acetylation (Low H3K14ac), based on data from Weiner et al. (2015), is plotted relative to the distance from the central dyad axis of the nucleosome (“dyad”). Vertical dotted lines indicate “Out” rotational settings. Nucleosomes overlapping with the MAG1 gene, which is deleted in the mag1Δ control strain, were excluded from this analysis. (B) Same as in part A, except for H3K36 trimethylation (H3K36me3). (C) Same as in part A, except for H3K79 monomethylation (H3K79me1).











