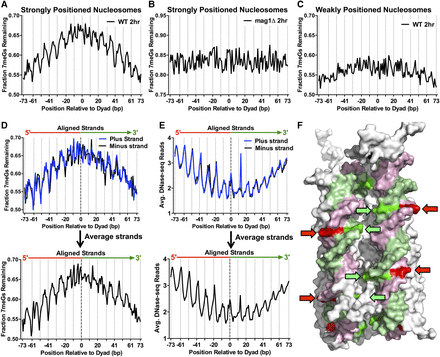
Repair of 7meG lesions is regulated by their rotational and translational setting within strongly positioned nucleosomes. (A) Fraction of 7meG lesions remaining following 2-h repair in WT relative to 0-h mag1Δ control among ∼10,000 strongly positioned nucleosomes (nucleosome score > 5) across the yeast genome is plotted relative to the distance from the central dyad axis of the nucleosome (“dyad”). Nucleosomes overlapping with the MAG1 gene, which is deleted in the mag1Δ control strain, were excluded from this analysis. Vertical dotted lines indicate “Out” rotational settings. (B) Translational and rotational setting among strongly positioned nucleosomes does not affect the fraction of 7meG lesions remaining following 2-h repair in a mag1Δ mutant relative to a 0-h control. (C) Same as in part A, except the fraction of 7meG lesions remaining is plotted among ∼7500 weakly positioned nucleosomes (nucleosome score < 1). (D) Effect of rotational setting on 7meG repair occurs primarily 5′ of the nucleosome dyad. Data are plotted similarly to part A, except the plus and minus strands are shown individually and aligned in a 5′ to 3′ orientation. Bottom panel: weighted strand average of fraction of 7meG lesions remaining for the aligned plus and minus strands. (E) The effect of rotational setting on DNase-seq read density, a measure of chromatin accessibility, also is more prominent 5′ of the nucleosome dyad. Bottom panel: average DNA-seq reads for the aligned plus and minus strands. (F) At “Out” rotational settings (indicated with arrows), the 5′ half of the nucleosomal DNA (pink/red) faces the solvent, while the 3′ half of the nucleosomal DNA (green) faces the other DNA gyre. Image was created using PyMOL (http://www.pymol.org/) to visualize PDB ID 1kx5.











