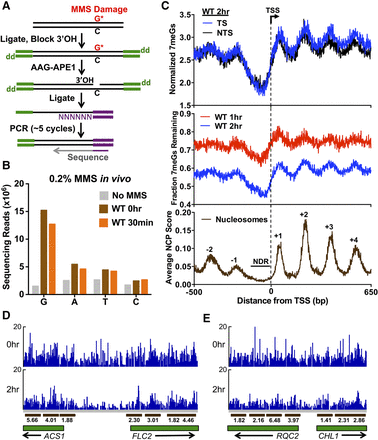
Genome-wide map of DNA alkylation damage and repair. (A) Experimental strategy for mapping N-methylpurine (NMP) lesions using NMP-seq. Isolated genomic DNA fragments are ligated to the trP1 adaptor (green), cleaved at NMP lesions with human alkyladenine glycosylase (AAG) and AP endonuclease (APE1), ligated to the biotinylated A adaptor (purple), purified, and sequenced. “dd” indicates dideoxy (i.e., 3′H). (B) Nucleotide distribution of lesion sites associated with NMP-seq sequencing reads. In MMS-treated samples, there is a significant enrichment of sequencing reads at “G”, and to a lesser extent, “A” nucleotides. (C) Nucleosome organization surrounding the transcription start sites (TSSs) of 5762 yeast genes modulates repair of 7meG lesions. Top panel depicts the average number of 7meG lesions per G nucleotide following 2-h repair in WT yeast strain. Middle panel depicts the fraction of 7meG lesions remaining following 1- or 2-h repair in WT relative to a matched 0-h mag1Δ control. Lower panel depicts the average nucleosome center positioning score (Brogaard et al. 2012). (D,E) Snapshot of the distribution of 7meG lesions at 0 h (mag1Δ) and following 2-h repair (WT) for the ACS1, FLC2, RQC2, and CHL1 genes. Dark brown bars indicate positioned nucleosomes, and the associated number indicates the nucleosome dyad score. Only well-positioned nucleosomes (NCP score ≥ 1) are depicted.











