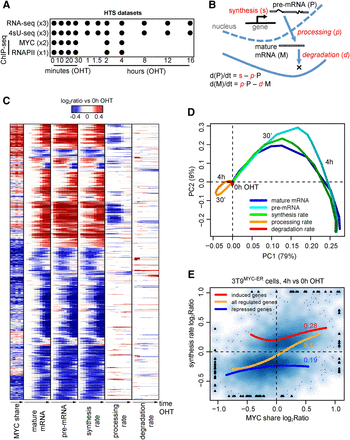
Dynamics of mRNA synthesis, processing, and degradation. (A) Study design indicating the collected HTS data, the time points of OHT treatment, and the number of replicates. In 4sU-seq samples, 4sU was added to the culture medium for 10 min prior to collection at every time point, to label and purify newly synthesized RNA. (B) Schematic representation of the kinetic rates (in red in the figure) of transcriptional regulation: immature and mature mRNA abundances, as well as synthesis rates, were derived directly from the experimental data, taking advantage of exonic and intronic reads in total and nascent RNA-seq data. Conversely, processing and degradation rates (in italic in the figure) were inferred from the integrated analysis of these data based on mathematical modeling. (C) Hierarchical clustering of the transcriptional response and change of MYC share for MYC-bound differentially expressed genes. Genes and time points are depicted in the rows and columns, respectively, and up- (red) or down- (blue) modulation is determined as the log2 ratio to the untreated condition. (D) Principal component analysis of the transcriptional response depicted in C; for each data type, subsequent time points of OHT treatment follow from light to darker shading; PC1 and PC2 are the first and second principal components, accounting for 80% and 9% of the explained variance, respectively. (E) Density scatter plot (darker colors for higher density) of the variation of MYC ChIP-seq signal within promoters (x-axis) vs. the corresponding change in synthesis rate (y-axis). The red, blue, and orange lines capture the trend for induced, repressed, and both (combined) set of genes, respectively. For the same set of genes, the Spearman correlation is reported.











