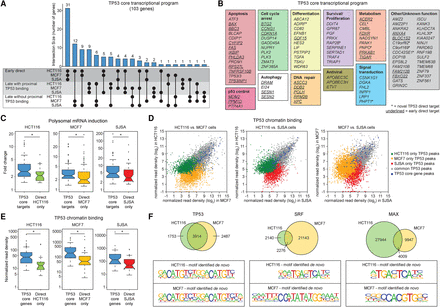
Identification of a core TP53 transcriptional program. (A) Upset plot showing conserved core TP53 target genes derived from the early direct and late target genes with and without proximal binding identified in HCT116, MCF7, and SJSA cells. (B) The core TP53 transcriptional program is composed of genes functioning in multiple effector pathways and genes of unknown function. Early direct target genes are underlined, previously unidentified targets are indicated with asterisks. (C) mRNA fold changes of core TP53 target genes versus cell-type–specific direct TP53 target genes (early direct and late targets with proximal binding combined). Statistics: Mann-Whitney U test, P < 0.01. (D) Scatter plots displaying normalized read densities for each TP53 ChIP-seq peak detected, for each pairwise comparison between cell lines. (E) Box and whisker plots showing normalized ChIP-seq read densities for TP53 peaks within 25 kb of the transcription start site of core TP53 target genes, compared to TP53 peaks associated with cell line-specific direct target genes. Statistics: Mann-Whitney U test, P < 0.01. (F) Venn diagrams comparing unique and overlapping (within a 100-bp window) transcription factor binding events in HCT116 versus MCF7 cells for TP53, SRF, and MAX. Bottom panels show the position weight matrices identified during de novo motif discovery for each cell line. See also Supplemental Figure S5 and Supplemental Files S6 and S7.











