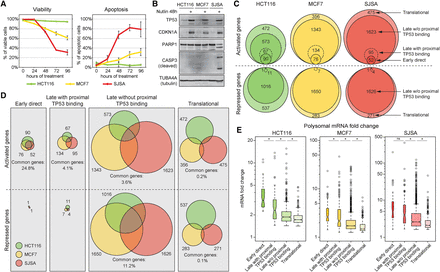
Cell-type–specific configurations of the TP53 signaling cascade. (A) Flow cytometric analysis of cellular viability (propidium iodide exclusion assay) and apoptosis (Annexin-V staining). Data points represent mean ± SD of three independent experiments. (B) Western blots showing TP53 and CDKN1A accumulation in HCT116, MCF7, and SJSA cells treated with 10 µM Nutlin-3 for 48 h. Only SJSA cells show cleavage of PARP and CASP3. (C,D) Venn diagrams illustrate fractions of genes up- or down-regulated in response to TP53 activation in each cell type. (E) Box plots showing fold change distributions of polysome-associated mRNAs across each cell line for the indicated categories. Statistics: Mann-Whitney U test, P < 0.01. See also Supplemental Figures S2 and S3, and Supplemental Files S1–S5.











