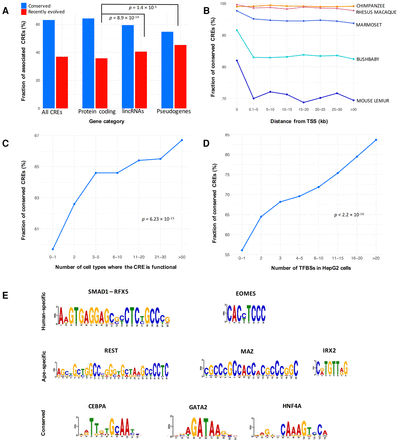
Genomic features associated with CRE conservation. (A) Fraction of conserved and recently evolved CREs associated with protein-coding genes, lincRNAs, and pseudogenes. (B) CRE conservation (y-axis) as a function of distance to the nearest gene start (quantiles on the x-axis). (C) CRE conservation (y-axis) as a function of cell-type specificity (quantiles on the x-axis) based on ENCODE data. (D) CRE conservation (y-axis) and number of ENCODE HepG2 TFBSs overlapping each CRE (quantiles on the x-axis). (E) Examples of enriched motifs in human-specific CREs, ape-specific CREs, and conserved CREs.











