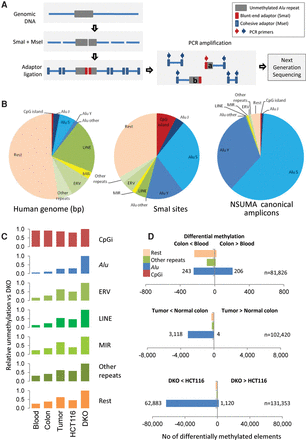
(A) Diagram illustrating the principle of Next-generation Sequencing of UnMethylated Alu repeats (NSUMA) technique. Genomic DNA is represented by a blue line, and the Alu repeat is represented by a gray box. DNA is digested with the restriction enzymes SmaI and MseI (impaired and insensitive to DNA methylation, respectively). The digested DNA fragments are ligated to two adapters with ends compatible with each one of the enzyme cuts. Primers complementary to the adapters are used to generate an amplified representation of the fragments. Amplicons generated from DNA fragments containing the SmaI adapter report unmethylated DNA and constitute the NSUMA canonical universe. Amplicons without the SmaI adapter (MseI-MseI) are also sequenced and used for the analysis of copy number variation. (B) Genetic element distribution according to the occupied fraction of the human genome, the content of SmaI sites, and their representation in the canonical NSUMA universe. (C) Relative unmethylation of different genetic elements in each one of the sample types analyzed by NSUMA in regard to the highly hypomethylated cell line DKO. CpG islands show the highest relative representation in all sample types as most of them are unmethylated, whereas Alu repeats show the lowest representation due to their heavy methylation. (D) Number of differentially methylated elements in the comparison between pairs of tissues (adjusted P-value <0.05 and |log2 FC| >1). Full data summary is reported in Supplemental Table S5, and detailed MA plots are shown in Supplemental Figure S13.











