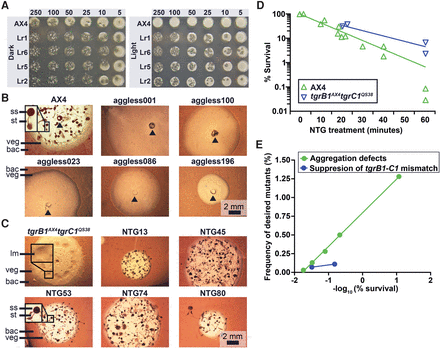
Genetic screens with chemical mutagenesis. (A) We compared the viability of the wild-type AX4 and the light-resistant (Lr#) mutants. We spotted 5 to 250 Dictyostelium cells on nutrient agar in association with bacteria. We incubated the plates in darkness (Dark) or under intense illumination (Light) for 4–5 d. Opaque circles are thick bacterial lawns (e.g., AX4, Light, 250) and dark plaques within the lawns are clearings that indicate amoebae growth. Whenever the amoebae cleared the bacterial lawns, starved and developed, a mass of fruiting bodies is evident as opaque spots and protrusions over the plaque background (e.g., Lr1, 250). (B,C) We inoculated the indicated Dictyostelium strains in association with bacteria (bac) on nutrient agar and incubated for 4–5 d. Cells (AX4 and NTG#) in the center of the plaque starved and developed into mature fruiting bodies with a sorus (ss) atop a stalk (st). Aggregation-less (aggless#) mutants did not form aggregates. tgrB1AX4tgrC1QS38 cells arrested at the loose mound (lm) stage. Vegetative (veg) Dictyostelium cells resided in the translucent periphery of the plaque. Insets are magnified (3×) for visual clarity. The black arrowheads indicate inoculation spots. Scale bar = 2 mm. (D) We measured the survival rate (%, y-axis) over time (minutes, x-axis) during mutagenesis of two strains, AX4 (n = 13) and tgrB1AX4tgrC1QS38 (n = 4). (E) We measured the frequency (%, y-axis) of mutants with the desired phenotypes and plotted against the negative logarithm of survival (%, x-axis) in the genetic screens for aggregation defects (n = 5) and for suppression of the tgrB1-tgrC1 mismatch phenotype (n = 2).











