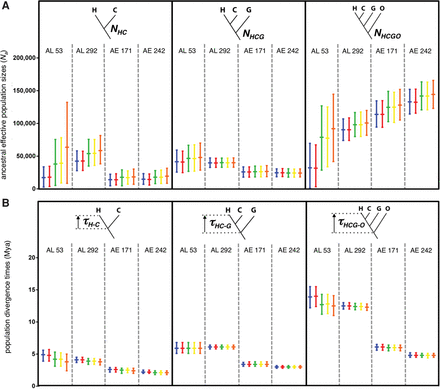
Bayesian estimation of ancestral effective population sizes and speciation times in the hominoids based on four phylogenomic data sets. (A) Inferred ancestral effective population sizes (Na) for the human–chimpanzee ancestor (NHC), human–chimpanzee–gorilla ancestor (NHCG), and human–chimpanzee–gorilla–orangutan ancestor (NHCGO). (B) Inferred speciation time in millions of years ago (Mya) between: human and chimpanzee lineages (τH-C), divergence leading to gorilla lineage (τHC-G), and divergence leading to orangutan lineage (τHCG-O). “AL 53” = 53 AL of Chen and Li (2001); “AL 292” = 292 AL; “AE 171” = 171 AE loci with each flanking sequence 500 bp from the adjacent AE; and “AE 242” = 242 AE loci with each flanking sequence 0 bp from the adjacent AE. Shown are posterior means and 95% credibility intervals for each parameter, which were estimated using five different sets of priors: P1 (blue), P2 (red), P3 (green), P4 (yellow), and P5 (orange; see Methods). Analyses for each parameter are contained within black boxes, and vertical dashed gray lines separate the results of each data set. Source data are in Supplemental Tables S6 through S13.











