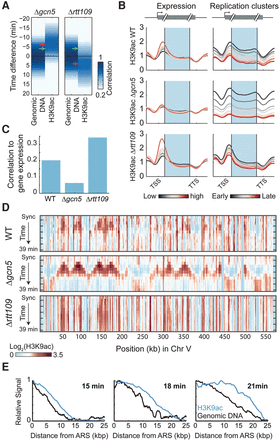
Replication-guided H3K9ac depends on Rtt109, while deletion of Gcn5 abolishes the expression-associated H3K9ac. (A) Prereplication H3K9ac depends on Rtt109. Same as Figure 2B for the indicated mutants. Arrows indicate the typical delay in the WT (green) or in the deletion strain (red). For H3K56ac in Δgcn5, see Supplemental Figure S9D. (B) Metagene pattern of H3K9ac in cells deleted of Gcn5 or Rtt109. Genes were separated into six groups based on their expression levels (left) or based on their replication clusters (right). The corresponding metagene patterns (cf. Fig. 1C) of H3K9ac in synchronized culture or in the middle of S phase (18 min) are shown, respectively. (C) Correlation between H3K9ac abundance and gene expression is lost in cells deleted of Gcn5. Correlations of gene expression to H3K9ac for the indicated strains at the synchronized time-point. Similar results were obtained at all other time-points. (D) The pattern of H3K9ac in wild type combines the replication- and transcription-associated patterns. H3K9ac (raw signal) in the indicated strains is plotted along chromosome V. (E) Spatial spreading of H3K9ac ahead of replication: All early ARS (<21 min in Yabuki et al. 2002) were aligned, and the average H3K9ac and genomic DNA signals were plotted over a 25-kb distance. Signal spreading is shown for three consecutive time-points in the gcn5-deleted strain and normalized by the synchronized time-point (Methods; see Supplemental Fig. S10 for H3K56ac).











