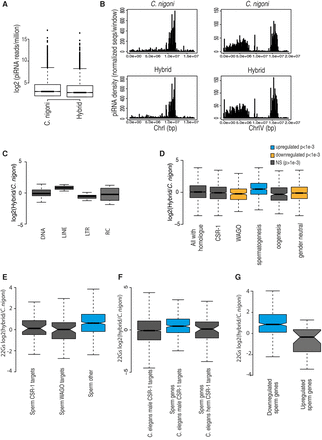
Comparison of expression of piRNAs and 22G RNAs between the males of ZZY10330 and C. nigoni (JU1421). (A) Comparison of read counts for piRNAs between C. nigoni males and the hybrid males (ZZY10330). (B) Distribution of piRNAs along the piRNA clusters on Chromosome I and Chromosome IV for C. nigoni (top) and the hybrid (bottom). The y-axis shows the number of unique piRNA sequences per million in each genomic window of 100 kbp. The x-axis shows position along the chromosome in base pairs. (C) Boxplot showing differences in 22G RNAs mapping antisense to different classes of TEs between hybrid males and C. nigoni males. Box shows interquartile range (IQ) with a line at the median, and the whiskers show the furthest point within 1.5 times the IQ range. (D) Boxplot showing differences between hybrid and C. nigoni in 22G RNAs mapping antisense to C. nigoni genes categorized by annotations from C. briggsae (CSR-1) or C. elegans (other categories). (E) Breakdown of spermatogenesis genes from D into CSR-1 and WAGO targets. (F) Differences between hybrid and C. nigoni either in all male-specific CSR-1 targets as defined in C. elegans or in the male-specific CSR-1 targets that overlap with spermatogenesis genes from D. (herm) Hermaphrodite. Boxplot parameters as in C. (G) Boxplot showing differences between hybrid and C. nigoni 22G RNAs mapping antisense to spermatogenesis genes found either up-regulated or down-regulated in hybrid males by mRNA-seq analysis (Fig. 3). Boxplot parameters as in C.











