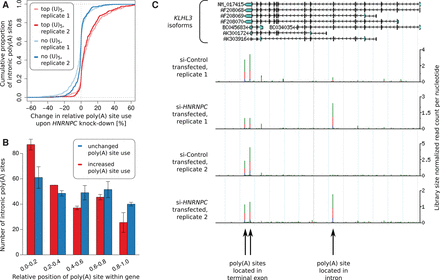
HNRNPC knock-down leads to increased usage of intronic poly(A) sites. (A) The change in the relative use of intronic poly(A) sites that did not contain any (U)5 within ±200 nt and of the top 250 intronic poly(A) sites according to the number of (U)5 motifs within ±50 nt around the cleavage site, upon HNRNPC knock-down. (B) Relative location within the gene of the top 250 most-derepressed intronic poly(A) sites that have HNRNPC binding motifs within −200 to +100 nt around their cleavage site and of the 250 intronic poly(A) sites that changed least upon HNRNPC knock-down. (C) Screenshot of the KLHL3 gene, in which intronic cleavage and polyadenylation was strongly increased upon HNRNPC knock-down.











