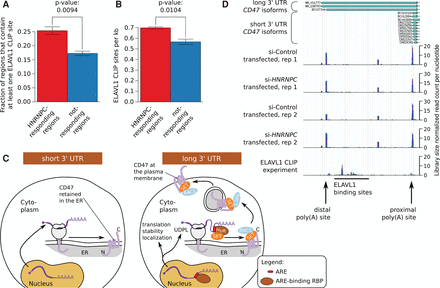
HNRNPC-responsive 3′ UTRs are enriched in ELAVL1 binding sites. (A) Fraction of HNRNPC-responding and not-responding 3′ UTR regions that contain one or more ELAVL1 CLIP sites. The P-value of the one-sided t-test is shown. (B) Density of ELAVL1 CLIP sites per kilobase (kb) in the 3′ UTR regions described above. The P-value of the one-sided t-test is shown. (C) Model of the impact of A/U-rich elements (ARE) in 3′ UTR regions on various aspects of mRNA fate (Berkovits and Mayr 2015). (D) Density of A-seq2 reads along the CD47 3′ UTR in cells, showing the increased use of the distal poly(A) site in si-HNRNPC compared with si-Control transfected cells. The density of ELAVL1 CLIP reads in this region is also shown.











