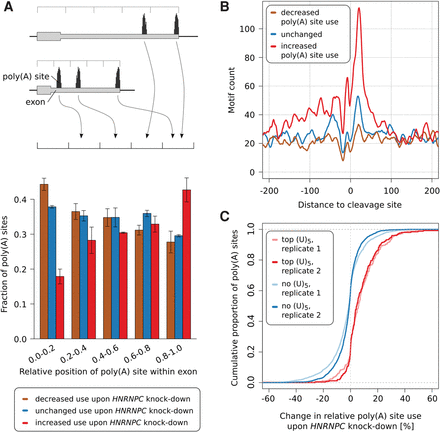
siRNA-mediated knock-down of HNRNPC leads to increased use of distal poly(A) sites. (A) Relative location of sites whose usage decreased (brown), did not change (blue) or increased (red) in response to HNRNPC knock-down within 3′ UTRs. We identified the 1000 poly(A) sites whose usage increased most, the 1000 whose usage decreased most, and the 1000 whose usage changed least upon HNRNPC knock-down; divided the associated terminal exons into five bins, each covering 20% of the exon's length; and computed the fraction of poly(A) sites that corresponded to each of the three categories within each position bin independently. Values represent means and SDs from the two replicate HNRNPC knock-down experiments. (B) Smoothened (±5 nt) density of nonoverlapping (U)5 tracts in the vicinity of sites with a consistent behavior (increased, unchanged, decreased use) in the two HNRNPC knock-down experiments. (C) Cumulative density function of the percentage change in usage of the 250 poly(A) sites with the highest number of (U)5 motifs within ±50 nt around their cleavage site (red) and of poly(A) sites that do not contain any (U)5 tract within ±200 nt (blue), upon HNRNPC knock-down.











