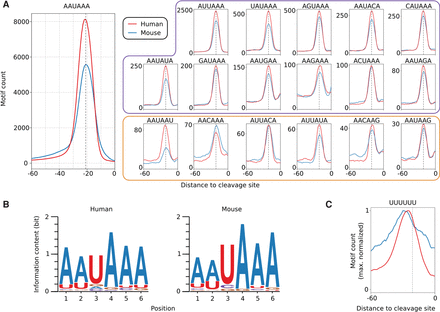
Hexamers with highly specific positioning upstream of human and mouse pre-mRNA 3′ end cleavage sites. (A) The frequency profiles of the 18 hexamers that showed the positional preference expected for poly(A) signals in both human and mouse. The known poly(A) signal, AAUAAA, had the highest frequency of occurrence (left). Apart from the 12 signals previously identified (AAUAAA and motifs with the purple frame) (Beaudoing et al. 2000), we have identified six additional motifs (orange frame) whose positional preference with respect to poly(A) sites suggests that they function as poly(A) signals and are conserved between human and mouse. (B) Sequence logos based on all occurrences of the entire set of poly(A) signals from the human (left) and mouse (right) atlas. (C) The (U)6 motif, which is also enriched upstream of pre-mRNA cleavage sites, has a broader frequency profile and peaks upstream of the poly(A) signals, which are precisely positioned 20–22 nt upstream of the pre-mRNA cleavage sites (indicated by the dashed, vertical line).











