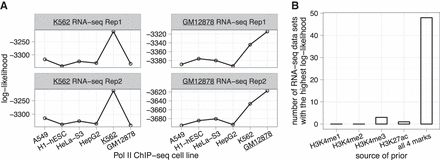
Informative priors for pRSEM can be derived from a broad range of data types. (A) Comparison of training set log-likelihoods based on isoforms partitioned by Pol II ChIP-seq peaks from six human cell lines. Log-likelihoods were computed by fitting pRSEM's Dirichlet-multinomial model to RNA-seq fragment counts of the partitioned training set isoforms. (B) Comparison of five sources of prior information by their effectiveness on 52 RNA-seq data sets of mouse hematopoietic cells. For each RNA-seq data set, all five sources were separately applied to fit the training set and the goodness of fit was assessed via the log-likelihood. The source resulting in the largest log-likelihood was considered to be the most effective. “All 4 marks” denotes a pRSEM partition model utilizing all four types of histone modification signals.











